Cell morphology analysis remains a cornerstone of biological research. From drug discovery screens to toxicology studies, researchers routinely examine thousands of microscopy images to classify cells by their phenotypic state. Yet manual annotation is slow, subjective, and increasingly impractical as imaging throughput accelerates.
The Scale of Manual Analysis
A typical high-content screening experiment generates tens of thousands of images containing millions of individual cells [1]. Classifying each cell as healthy, stressed, dividing, or dying requires trained experts who can identify subtle morphological features: membrane integrity, nuclear condensation, cytoplasmic organization, and cell shape changes.
Manual classification rates average 100-300 cells per hour for experienced analysts. At this pace, a modest experiment with 50,000 cells requires 150-500 hours of human labor. The bottleneck is not image acquisition but interpretation.
Recognizing Cell States
Cells display characteristic morphological signatures across phenotypic states:
Healthy cells typically appear rounded or spread (depending on cell type), with intact membranes, uniformly distributed chromatin, and organized cytoplasm.
Elongated cells often indicate stress responses, migration, or differentiation. The cytoskeleton reorganizes, extending the cell body along one axis.
Apoptotic cells exhibit membrane blebbing, chromatin condensation, nuclear fragmentation, and eventual cell shrinkage. These morphological changes follow a predictable sequence during programmed cell death [2].
Distinguishing these states requires integrating multiple visual features, a task humans perform intuitively but inconsistently across long annotation sessions.
Deep Learning Transforms Cell Analysis
Recent advances in convolutional neural networks have enabled automated cell segmentation and classification at unprecedented accuracy. The TissueNet dataset, containing over one million manually labeled cells, demonstrated that deep learning models can achieve human-level performance in identifying cell boundaries across diverse tissue types [3].
These models learn hierarchical feature representations: edges and textures at early layers, cell shapes and organelle patterns at deeper layers. Once trained, they process images in milliseconds rather than minutes.
The Ambiguity Challenge
Not every cell fits neatly into predefined categories. Transitional states, imaging artifacts, overlapping cells, and rare phenotypes create ambiguous cases where even experts disagree. Binary classification systems fail here, either forcing incorrect labels or discarding valuable data.
Intelligent classification systems address this by quantifying prediction confidence. Cells with high-confidence predictions proceed automatically; ambiguous cases are flagged for expert review. This hybrid approach preserves human judgment where it matters while automating routine classification.
AI Agents for Phenotype Classification
An AI agent configured for cell phenotype analysis can automate the complete workflow:
Image preprocessing: Normalize illumination, correct for background fluorescence, and segment individual cells from dense populations.
Feature extraction: Quantify morphological parameters including area, perimeter, circularity, aspect ratio, and texture features that distinguish phenotypic states.
Classification with confidence: Assign phenotype labels while flagging low-confidence predictions for manual review.
Result aggregation: Compile statistics across experimental conditions, generate visualizations, and identify outlier populations.
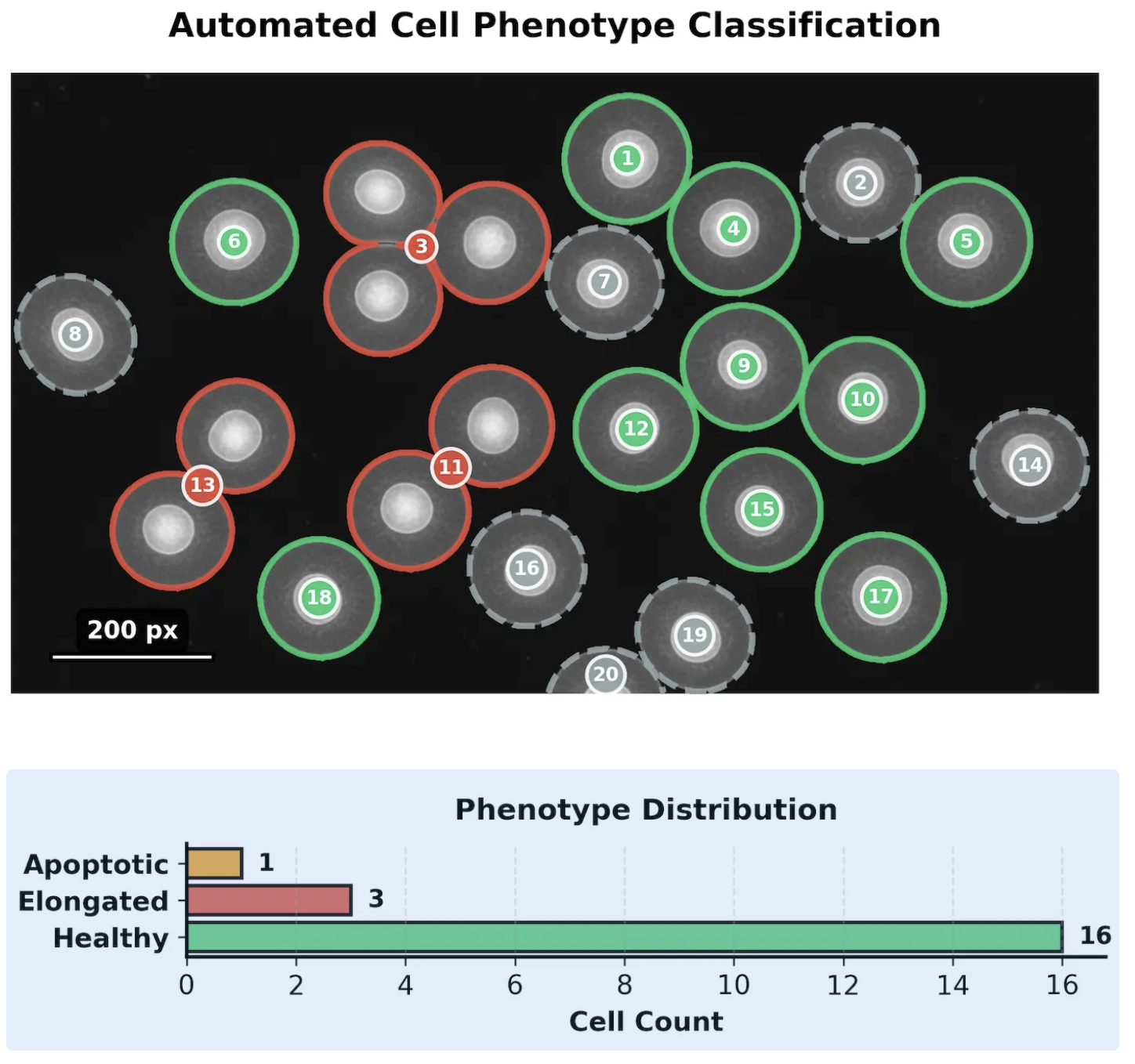
The agent reduces analysis time from hours to seconds while maintaining quality through intelligent uncertainty quantification.
Practical Impact
For drug discovery, faster phenotype classification accelerates compound screening. Toxicity signals emerge earlier. Dose-response curves complete in days rather than weeks.
For basic research, automated analysis enables experiments previously impractical due to scale. Time-lapse studies tracking thousands of cells through division, differentiation, or death become routine.
For clinical applications, standardized classification reduces inter-observer variability that complicates diagnostic interpretation.
The Path Forward
As imaging technologies generate ever-larger datasets, manual analysis becomes the limiting factor in cell biology research. AI agents that combine deep learning classification with intelligent uncertainty handling offer a path to scale, automating routine decisions while preserving human expertise for genuinely ambiguous cases.
References
[1] Chandrashekar DS, et al. "UALCAN: An update to the integrated cancer data analysis platform." Neoplasia. 2022;25:18-27. https://doi.org/10.1016/j.neo.2022.01.001
[2] Klionsky DJ, et al. "Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition)." Autophagy. 2021;17(1):1-382. https://doi.org/10.1080/15548627.2020.1797280
[3] Greenwald NF, et al. "Whole-cell segmentation of tissue images with human-level performance using large-scale data annotation and deep learning." Nature Biotechnology. 2022;40(4):555-565. https://doi.org/10.1038/s41587-021-01094-0